A family affair: understanding a parasite surface protein family
A family affair: understanding a parasite surface protein family
Why do parasites often have protein families?
Human blood is an excellent place for a parasite to live as it is full of nutrients that can support parasite growth. But the molecules and cells of the immune system make blood a hostile environment, as they recognise parasite molecules and destroy parasites.
Parasites which survive in blood often make use of large surface protein families to interact with their human host. The presence of a diverse family of surface proteins with similar function allows the parasite to switch which protein it uses to perform each particular role. When the human immune system learns to recognise one member of the family, the parasite can switch to use another in the process of antigenic variation.
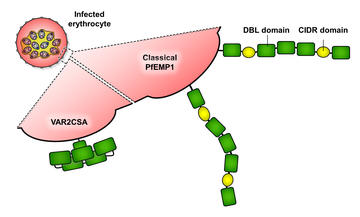
The parasites that cause malaria dwell within human red blood cells. This is a winning strategy as it allows them to hide from detection. But they still need surface molecules. In particular, red blood cells circulating around the body pass through narrow slits in the human spleen. Parasite-laden blood cells will be filtered away and destroyed. For this reason, the parasite sends molecules from the PfEMP1 surface protein family to the red cell surface. These PfEMP1 are modular, made from a series of individual structural units and these individual domains can bind to different human receptors, such as EPCR, CD36 and ICAM-1. This causes infected blood cells to stick to tissue and blood vessel surfaces, saving them from clearance in the spleen.
In many ways, the PfEMP1 are the classic parasite surface protein family. There are many PfEMP1 available to each parasite strain and they live under conflicting evolutionary pressures. On one hand, they diversify to provide an antigenically distinct set of molecules to aid parasite survival. On the other hand, they retain the ability to bind to specific human receptors. How do they balance these challenges at a molecular level?
In a series of studies, mostly in collaboration with Thomas Lavstsen, Anja Jensen, Louise Turner and colleagues at the University of Copenhagen, we have tackled these questions. What have we discovered?
How far does family similarity go?
To work out how PfEMP1 proteins balance the need to diversify with the need to retain capacity to bind their human receptors, we used structural methods. We revealed the molecular features needed for binding to the three major human receptors to which PfEMP1 stick; EPCR, CD36 and ICAM-1. By mapping sequence data onto these structures, we then identified which regions are variable and which are conserved. So, how much variation is possible in PfEMP1 domains which bind to a single ligand?
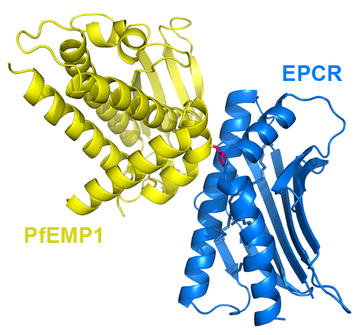
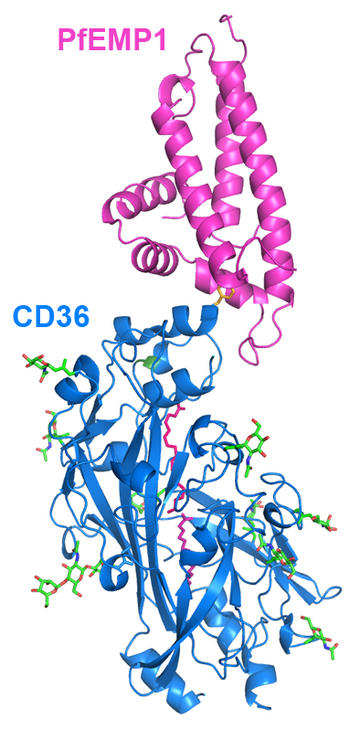
Our first structure was that of a PfEMP1 domain bound to EPCR. Here, the binding site centres around a hydrophobic bump, surrounded by a hydrophilic surface. There is remarkable sequence variation across this surface, with many differences even in the amino acids which directly contact EPCR. Despite this, the chemistry and shape of the EPCR-binding surface is retained. A similar story emerged from our structure of a PfEMP1 domain bound to CD36. Here again we see a binding surface of variable sequence but conserved shape. Is this typical?
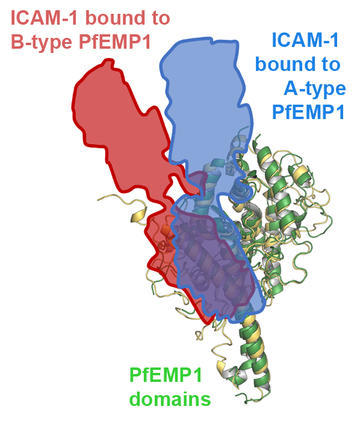
There are two different groups of PfEMP1 domains that bind to ICAM-1. One of these, known as the B-type ICAM-1 binders, also shows major sequence diversity, even in the regions which bind to ICAM-1. In contrast, A-type ICAM-1 binders were surprisingly different, as they have a region of nearly total sequence conservation at the heart of their ICAM-1 binding site. The reasons for this are unknown, but the A-type ICAM-1 binders are clearly the rare identical twins of the PfEMP1 world, rather than displaying the usual deep diversity characteristic of the family.
These studies reveal a remarkable level of sequence variation in domains which can still bind to the same human receptor. Nevertheless, underneath this surface variation the family resemblance can still be seen. Domains retain the same shape and chemistry to allow them to continue to bind to the same receptor. While they may diversify to allow them to seem different to antibodies, they retain their basic similarity, allowing them to perform the roles that help keep the parasite alive.
Sticking to the business part of human receptors
The malaria parasite is an ancient human foe with whom we have been co-evolving for millennia. The presence of malaria has driven changes in the human genome in areas of high malaria burden, with mutations which reduce our risk of getting malaria proving beneficial to survival. So why have CD36, EPCR and ICAM-1 not mutated to stop PfEMP1 from binding?
Our structures reveal that PfEMP1 often bind to the business part of human receptors. The natural function of EPCR is to bind a signalling molecule, human protein C and both PfEMP1 and protein C bind to the same site on EPCR. Similarly, the natural function of CD36 is to bind to lipid rich lipoprotein particles to help human cells to acquire this nutrient. Both PfEMP1 and lipoprotein particles bind to the same site on CD36.
This is a cunning strategy for the parasite. Functionally important sites on human receptors cannot easily be changed without losing their important role in the human body. By binding to such sites, the parasite reduces the chance of a human mutation occurring which makes the parasite unable to bind.
But this may have serious consequences for the host. If PfEMP1 proteins bind to functionally important sites, then they can interfere with function. Indeed, PfEMP1 binding appears to prevent protein C from triggering signalling through EPCR. This may be how it causes inflammation and contributes to severe malaria.
Different parts working as a whole?
Each PfEMP1 contains multiple domains. These are often thought to be individual modules with their own binding properties. But neighbouring modules within the same PfEMP1 can cooperate, working together to help the parasite.
Our first example of PfEMP1 in which neighbouring domains act together is those that contain an A-type ICAM-1 binding domain. All of these proteins also contain a neighbouring EPCR-binding domain. These two domains can act at the same time, causing parasites to stick more tightly to surfaces which contain both EPCR and ICAM-1.
These dual binding phenotypes have consequences for PfEMP1 organisation. For both domains to simultaneously bind both ligands, the domains have to have the correct spatial arrangement to match the arrangement of the receptors to which they bind. This may explain why there are two types of ICAM-1 binding domain. As well as the dual ICAM-1 and EPCR-binding PfEMP1, there are also PfEMP1 that contain neighbouring CD36-binding domain and B-type ICAM-1 binding domains. Our structures show that ICAM-1 binds with a different angle to the A- and the B-type ICAM-1 binding domains. Perhaps this is because EPCR and CD36 protrude to different degrees from the membrane, making different geometries of ICAM-1 binding necessary to allow neighbouring PfEMP1 domain to bind to these different ligands. The constraint to allow beneficial dual binding appears to have affected the evolution of these proteins in multiple ways.
The dangerous members of the family
Like any family, not all PfEMP1 are all equal in temper. Some are associated with mild episodes of malaria while others are more commonly used by parasites causing episodes of severe malaria. In one case, the link is very clear. The expression of a particular PfEMP1, known as VAR2CSA is very tightly linked to the development of pregnancy-associated malaria, where it tethers infected erythrocytes within the placenta. In other cases, the association is less absolute. But PfEMP1 which bind to EPCR are more likely to be found in children suffering from severe malaria while those with dual ICAM-1 and EPCR-binding phenotypes are more likely to be used by parasites causing cerebral malaria. Whether these interactions can be prevented by an effective vaccine which works against all parasite strains remains to be seen. But the diversity shown, even in PfEMP1 which bind to a single receptor, will bring significant challenges.
A depth of complexity
These studies have brought PfEMP1 into close focus for the first time, perhaps providing the most detailed molecular insight into any parasite surface protein family and its interactions with its host receptors. They reveal a highly diverse set of proteins, which maintain surfaces of conserved chemistry to allow them to bind to specific host receptors. They show that PfEMP1 often bind to functionally important and invariant sites on these receptors. We see that, while PfEMP1 are modular, with individual domains generally binding to individual receptors, these modules often work together and their evolution is influenced by their neighbours. Finally, we see that PfEMP1 with different binding partners are associated with different disease outcomes. With so many different PfEMP1 domains with no known host binding partner, it is likely that we have just scratched the surface of their deep complexity.
To find out more:
Lennartz, F., Smith, C., Craig, A.G. and Higgins, M.K. (2019) Structural insights into diverse modes of ICAM-1 binding by Plasmodium falciparum-infected erythrocytes. Proc. Natl. Acad. Sci. USA 11620124-20134
Lennartz, F., Adams, Y., Bengtsson, A., Olsen, R.W., Turner, L., Ndam, N.T., Ecklu-Mensah, G., Moussiliou, A., Ofori, M.F., Gamain, B., Lusingu, J.P., Petersen, J.E.V, Wang, C.W., Nunes-Silva, S., Jespersen, J.S., Lau, C.K.W., Theander, T.G., Lavstsen, T., Hviid, L., Higgins, M.K.*, Jensen, A.T.R * (2017) Structure-guided identification of a family of dual receptor binding PfEMP1 that are associated with cerebral malaria. Cell Host and Microbe 21 403-414
Hsieh, F., Turner, L., Bolla, J.L., Robinson, C.V., Lavstsen, T. and Higgins, M.K. (2016) The structural basis for CD36 binding by the malaria parasite. Nature Comm 7 12837
Lau, C.K.Y, Turner, L., Jespersen, J.S., Lowe, E.D., Petersen, B., Wang, C.W., Petersen, J.E.V., Lusingu, J., Theander, T.G., Lavstsen, T.* and Higgins, M.K.* (2015) Structural conservation despite huge sequence diversity allows EPCR binding by the PfEMP1 family implicated in severe childhood malaria. Cell Host and Microbe 17 118-129
Turner L., Lavstsen T., Berger S.S., Wang C.W., Petersen J.E.V., Avril M., Brazier A.J., Freeth J., Jespersen J.S., Nielsen M.A., Magistrado P., Lusingu J., Smith J.D., Higgins M.K., Theander T.G. (2013) Severe malaria is associated with parasite binding to endothelial protein C receptor. Nature 498 502-5.


